Navigation
What is iNuLoC
iNuLoC is a platform developed for understanding the nuclear location codes in proteins. We developed a deep learning based model to predict the nuclear location proteins, discovered complex biological rules to decipher protein nuclear location potential. iNuLoC provided the prediction results of pNuLoC and integrated several well-known database including UniProt, NLSdb, SeqNLS, ValidNESs and NESbase to display known and candidate NLSs/NESs in the proteins, which might provide helpful information for the research of protein nuclear location.
About predicting performance
ROC for cross validation and test validation
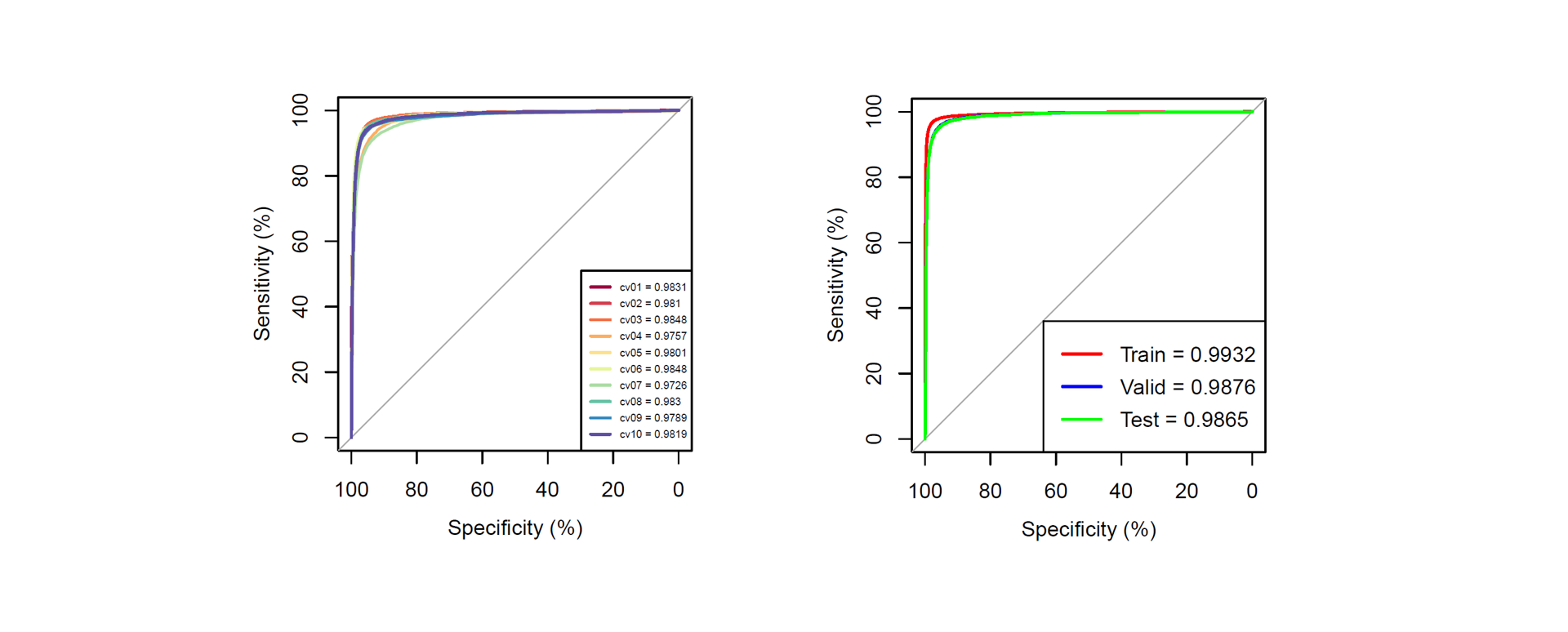
Threshold selection and performance
| Threshold | Score | Specificity | Sensitivity | Precision | Recall | Accuracy | MCC |
|---|---|---|---|---|---|---|---|
| High | 0.737 | 0.990 | 0.941 | 0.970 | 0.941 | 0.977 | 0.940 |
| Medium | 0.476 | 0.950 | 0.975 | 0.872 | 0.975 | 0.957 | 0.893 |
| Low | 0.328 | 0.900 | 0.982 | 0.774 | 0.982 | 0.921 | 0.822 |
Search & Browse
Quick Search
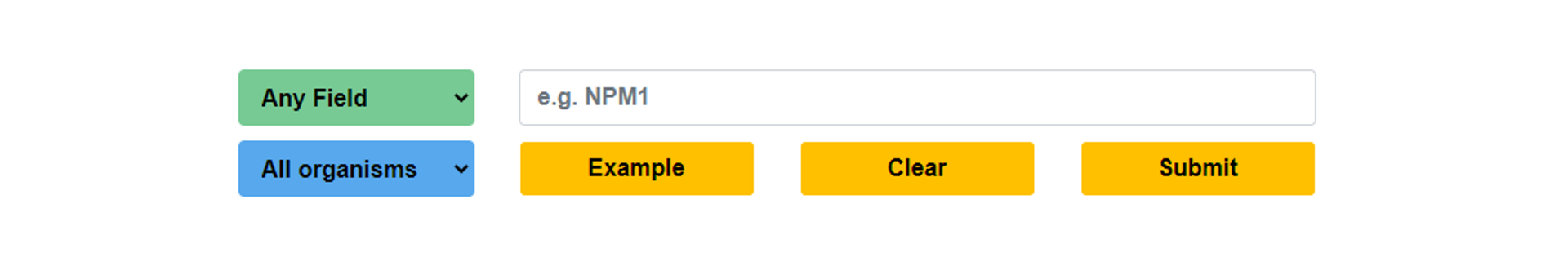
In the home page of iNuLoC, users can specify one field or all the fields to perform a quick search with the keyword(s) in the field(s). Users can simply click example and submit buttons to perform an example search.
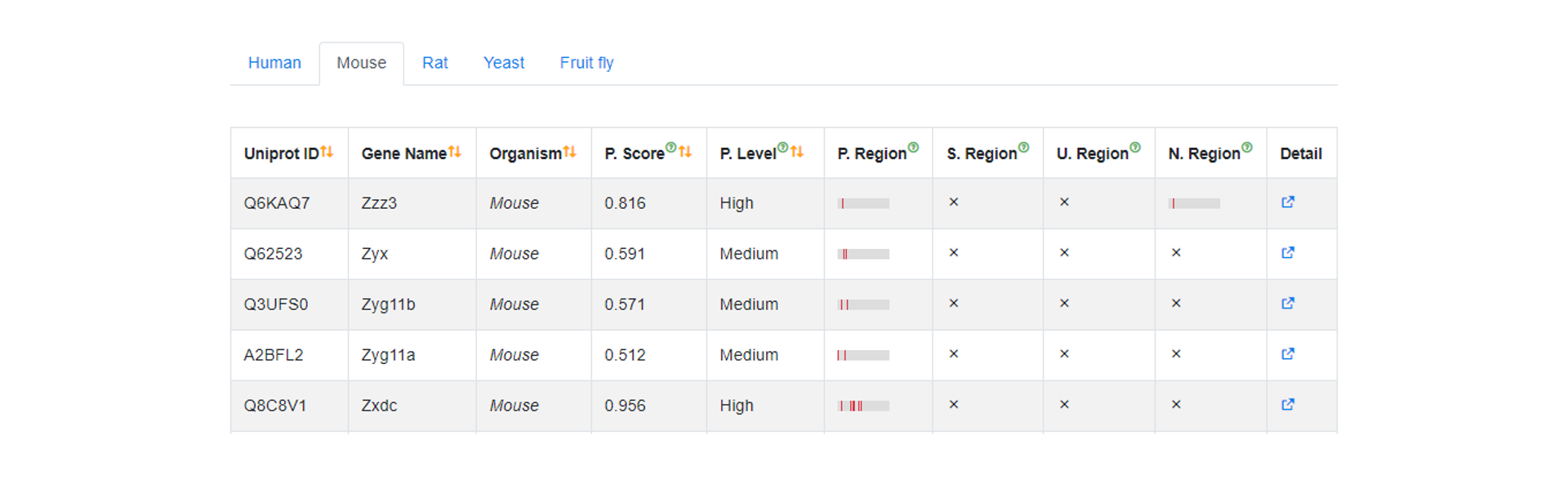
Browse
In the Browse page, users can click one term to search the data of specified organism in iNuLoC. Users can click the Detail buttom to display the term of interest.
pNuLoC webserver
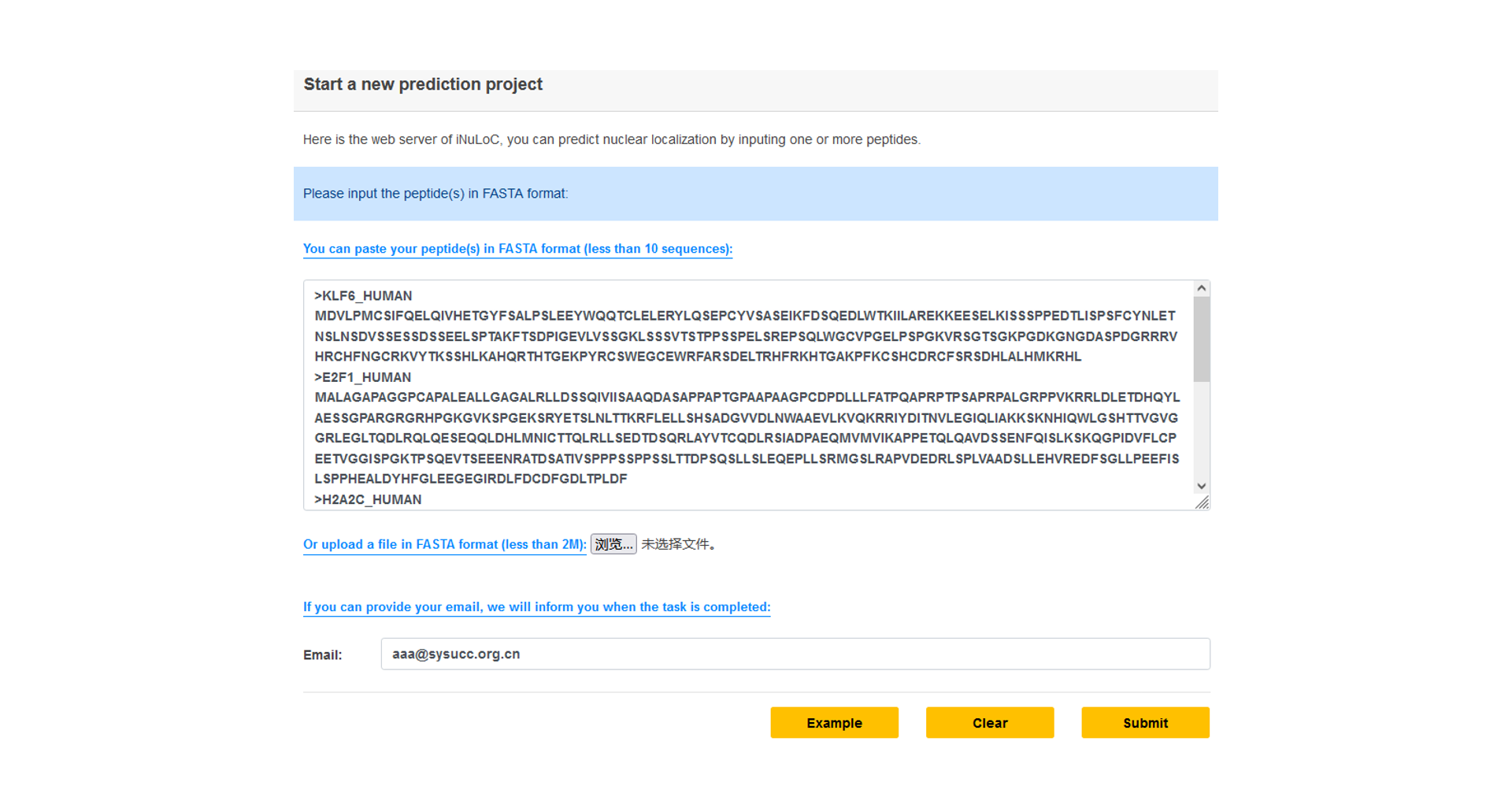
Submit a new task
Step1: Paste your FASTA format sequence into the input textarea, or you can click the example button to run the default sequence.
Step2: If you provide email address, we will send you the task id by email, or you can see your tasks at "All submitted projects".
Step3: Click Submit.
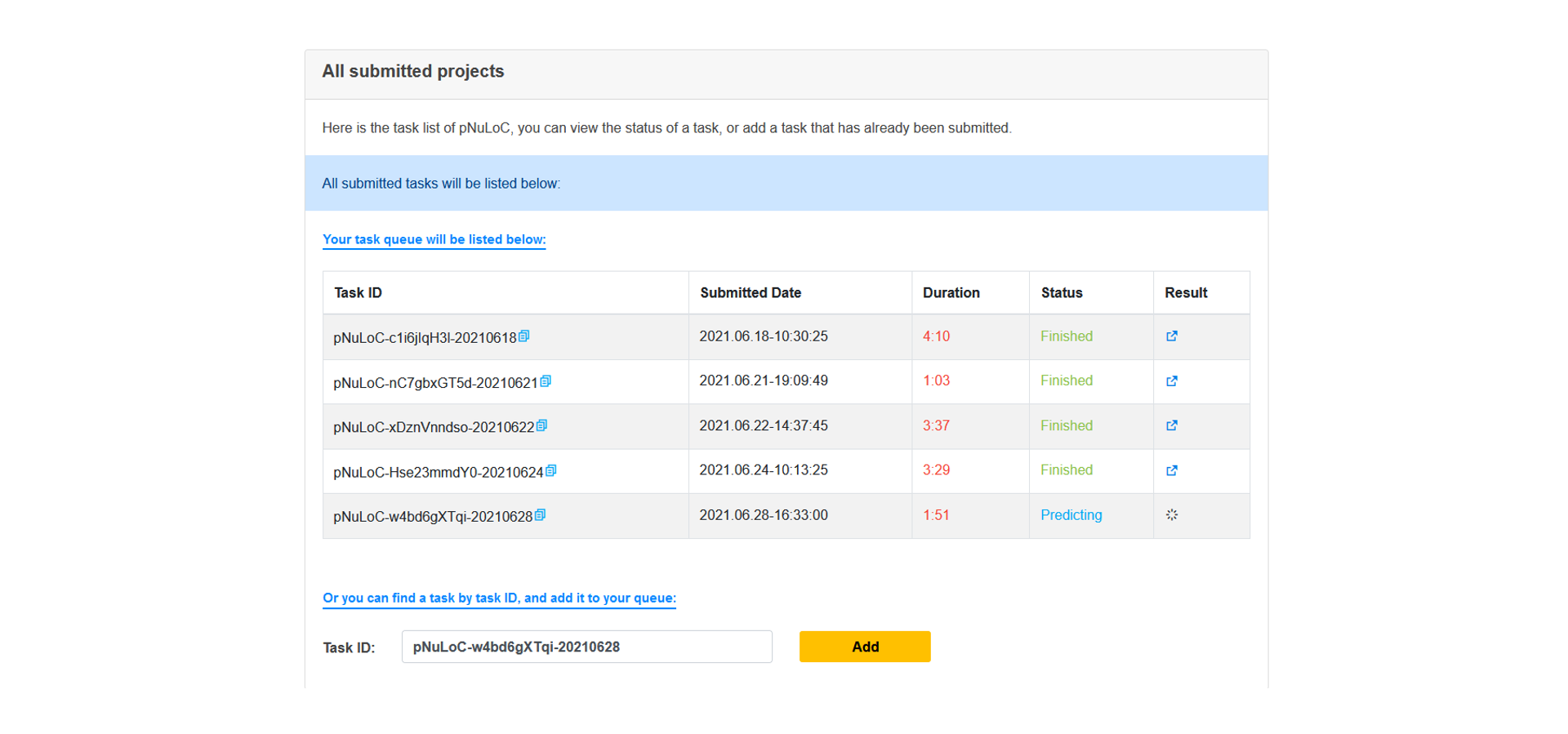
Query a submitted task
All submitted tasks will be listed below, Users can view the status of a task, or add a task that has already been submitted.
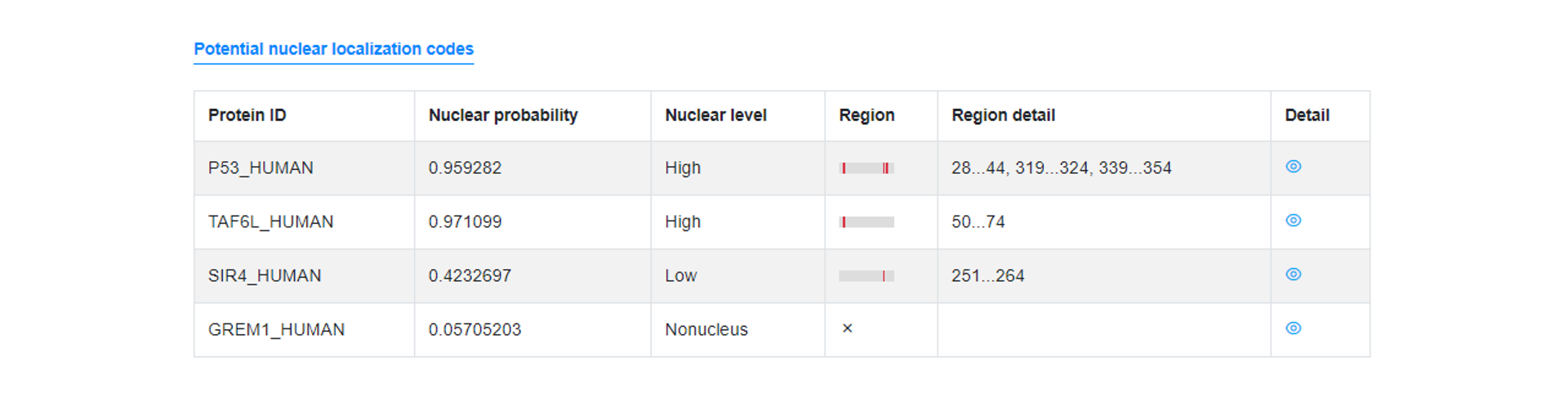
Output result explanation
Potential nuclear location probability
This table contains five column, including FASTA title ID, Nuclear location probability, Confidence level, Possible regions.
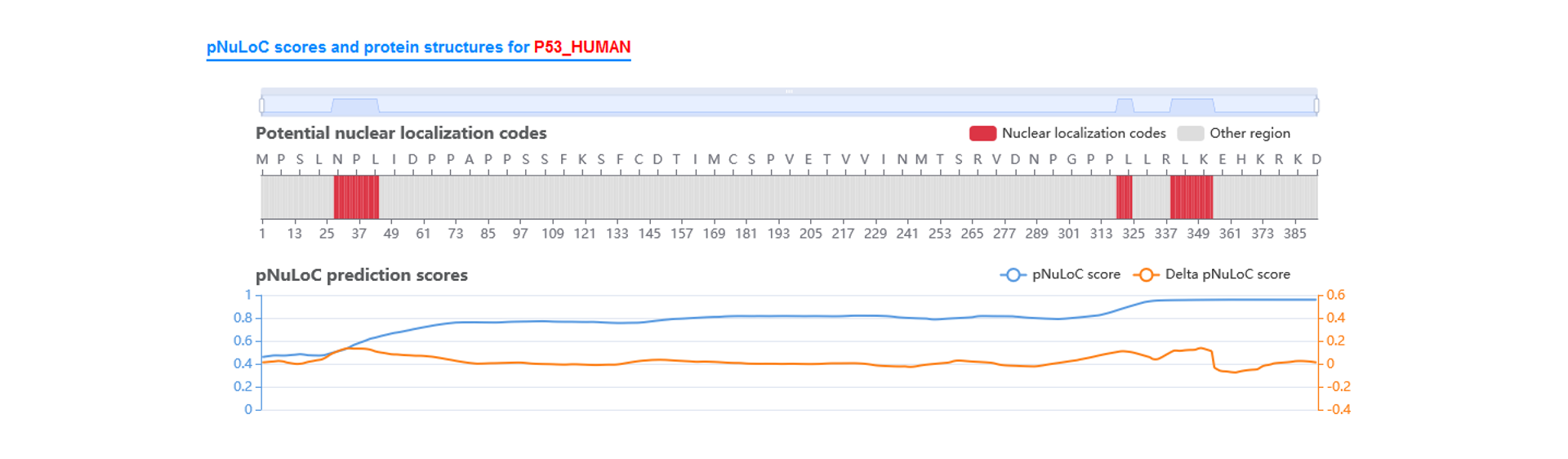
Potential nuclear location regions
Based on pNuLoC, we calculated the nuclear location probability trajectory score for each site.
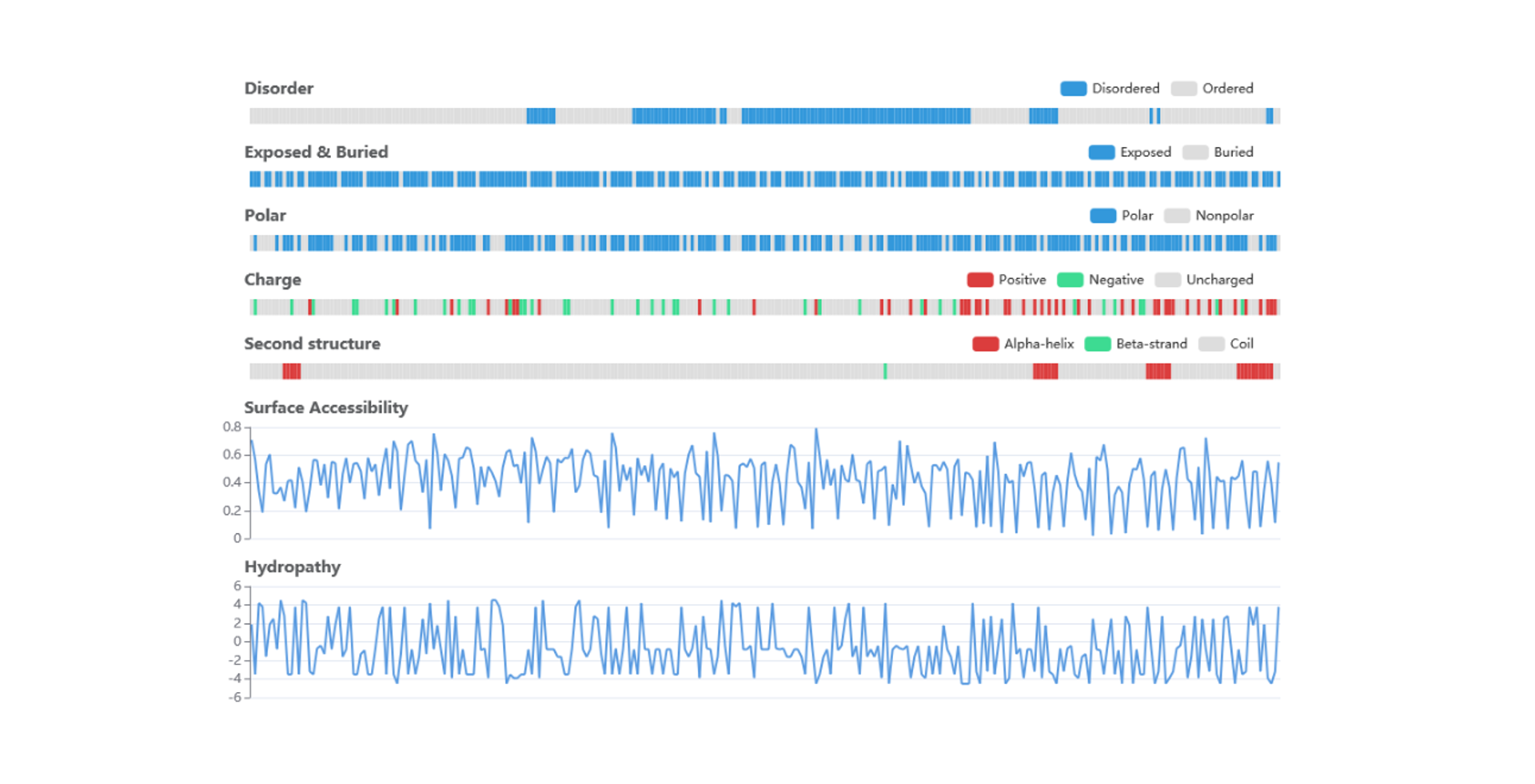
Sequence and structure properties
The disorder values are calculated by IUPred. The surface accessibility and secondary structure information are predicted by NetSurfP. The polar, charge and hydropathy information were retrived from AAindex.